|

|
| Paradigm Boa |
Here we will consider a test cross with a Paradigm that can produce Paraglows,
but this time using a Salmon-hypo het Sharp-albino (also known as a double-het Sharp-sunglow) rather than a
Sharp-sunglow.
The analysis begins in the same manner as before.
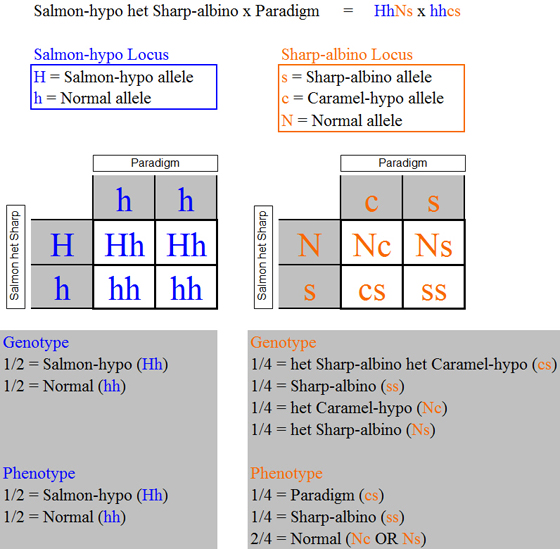
As in the previous example, at the Salmon-hypo locus each possible phenotype
is associated with a unique genotype. However, this is not the case at the Sharp-albino
locus where there are four possible genotypes but only three possible phenotypes that can result. The standard approach is to construct the tables based on the genotypes and their frequencies as shown
below.
|
| Click on the image to view full size. |
Note that in the tables we have
used some mixed terminology, with some of the names for the genotypes technically being phenotypes; such terminology substitutions
are acceptable so long as the phenotype corresponds to a unique genotype at least via definition. For example, Paradigm is technically a phenotype, whereas the associated genotype is het Sharp-albino het
Caramel-hypo. Because the phenotype and genotype are uniquely associated with
each other, there is no confusion in using the name Paradigm for the genotype. The
true genotype names often can get quite lengthy (e.g., Salmon-hypo het Sharp-albino het Caramel-hypo), and so it is a simple
matter of convenience to substitute the uniquely associated phenotype name (Paraglow in this case). Always carrying the genotypes through the tables in the form of their allelic representations (e.g., Paraglow
= Hhcs) eliminates any possible confusion.
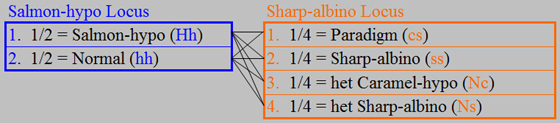
We now have all of the expected
genotypes and frequencies for the offspring, but there is one more step that we need to complete. We must add another column to the table showing the offspring
phenotypes to tell us which genotypes will be visually distinguishable in the offspring and the expected frequency of each
phenotype.

|
| Click on the image to view full size. |
Each of the Paraglow,
Paradigm, Sharp-sunglow, and Sharp-albino phenotypes is associated with its unique genotype, so we will know the genotypes
of these offspring by their physical appearance, and each will occur at an expected frequency of 1/8. However, the phenotype Normal offspring, which are expected to occur at a frequency of 2/8 (1/8 + 1/8) could have either of
two genotypes: het Sharp-albino or het Caramel-hypo, and these two genotypes
are expected to occur at equal frequencies. In other words, the normal-appearing
offspring are either het Sharp-albino or het Caramel-hypo, the odds are 50/50 as to which for any particular individual, and
you can’t know which for any particular individual until you grow it up and breed it.
This should be sounding familiar by now, as it is our definition of a ParaHet!
So, we now know that the phenotype Normal (i.e. normal-appearing)
offspring are ParaHets, and will occur at an expected frequency of 2/8. Similarly,
our definition of the ParaHet genotype allows us to say that the phenotype Salmon-hypo offspring will be genotype Salmon-hypo
ParaHet, and will occur at an expected frequency of 2/8.
|
| Click on the image to view full size. |
This method of
constructing the tables based on the genotypes and their frequencies, filling in the column of offspring phenotypes based
on the offspring genotypes, and finally evaluating whether each offspring phenotype is associated with a unique genotype or
if there is one or more expected offspring phenotype with multiple possible genotypes (i.e., if there are expected offspring
whose genotypes will not be known based on their appearance) will always work, and provides all of the details of the expected
genotypes, phenotypes, and their expected frequencies.
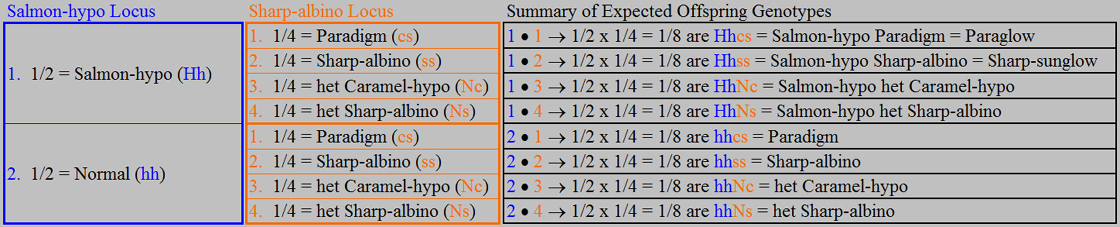
However, there is a shortcut that
can get us to the answers we are usually most interested in, and eliminates the need for the phenotype evaluation at the end. We begin with the same set of Punnett squares and genotype/phenotype breakdown for
each locus as before.
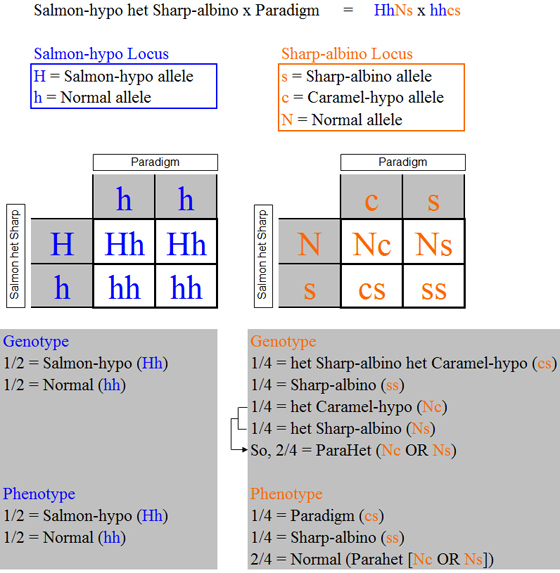
As noted previously, each phenotype
at the Salmon-hypo locus is associated with a unique genotype, but at the Sharp-albino locus there are three possible phenotypes
and four possible genotypes. Rather than constructing the tables based on the
genotypes as before using the four Sharp-albino locus genotypes that each occurs at a frequency of 1/4, we observe that the
Nc and Ns genotypes can be combined using our ParaHet genotype definition. To
say that there are an expected 1/4 het Caramel-hypos and 1/4 het Sharp-albinos is equivalent to saying that there are an expected
2/4 ParaHets as shown above (or, to say that there are an expected 2/4 phenotype Normal offspring, half of which on average
will be het Caramel-hypos and half of which on average will be het Sharp-albinos, is equivalent to saying that there are an
expected 2/4 ParaHets).
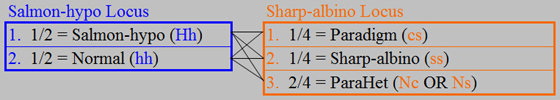
This use of the ParaHet genotype definition provides us with a unique genotype for
each phenotype. At the Sharp-albino locus, all normals (i.e., any offspring not
Paradigm or Sharp-albino) are ParaHets. Another way of looking at it is to say
that we are constructing the tables based on the phenotypes and carrying the needed genotype information along with each phenotype.
Constructing our tables on this basis and carrying the allelic representations
of the genotypes (e.g., Paradigm = “cs”) along in parentheses as before provides a somewhat simpler and more direct
result as shown below.

|
| Click on the image to view full size. |
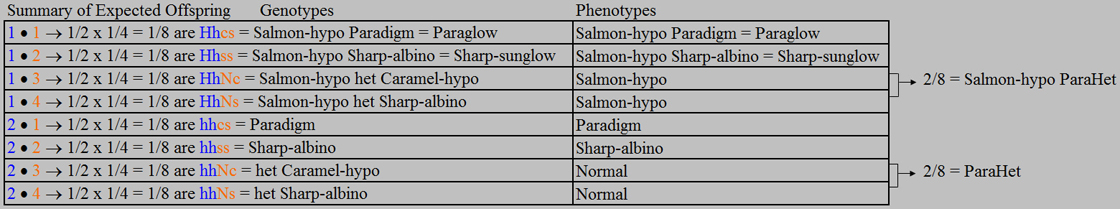
Technically, this expected offspring
summary does not directly provide us with every detail about the genotypes, phenotypes, and their frequencies. For example, we must know the definition of a ParaHet, and we must know that a Salmon-hypo
ParaHet will look like any other Salmon-hypo that is not also het for Caramel-hypo or Sharp-albino (i.e., not also ParaHet). Of course, this is information that we need to know anyway if we are going to be breeding
Paradigm boas and related morphs. All of the details about the genotypes, phenotypes and frequencies can easily be extracted
from this table. Once one is comfortable with the terminology and definitions, this shortcut method is very effective.
One can always resort to the standard
approach above if the shortcut method proves too confusing. The key distinction
to remember is that the genotype refers to the actual combinations of alleles at the loci being evaluated, and the phenotype
(at least for the purposes of visual morphs in boas) refers to a boa’s physical appearance. In the remainder of the example test cross analyses, we will use the shortcut method presented above. We will also not maintain strict distinction between genotype and phenotype names,
and in particular may at times use the terms Paradigm and ParaHet to describe both genotype and phenotype where applicable.
Here we offer one final
observation about the outcome of this test cross. Whereas the Sharp-sunglow to
Paradigm test cross analysis showed that on average 1/4 of the offspring are expected to be Paraglow, with this test cross
of a Salmon-hypo het Sharp-albino (double-het Sharp-sunglow) to Paradigm we find that only 1/8 of the offspring are expected
to be Paraglow. These types of comparisons are useful in assessing a boa’s
relative potential breeding value, planning boa production with an eye toward desired future breeding trials, and helping
a breeder to strike a responsible balance between the value of outcrossing and the probability of producing a desired boa
morph.
Continue to "6. Paradigm x Motley het Sharp-albino"
Back to "4. Multiple Traits"
Back to the "Genetics and Paradigms" Page
|

